By G. Kago, Post Doctoral Fellow, Molecular Biosciences
Welcome to the LaMontagne Center for Infectious Disease Blog!
Here we will feature posts for both scientists and non-scientists regarding infectious disease research at the LaMontagne Center for Infectious Disease. Some posts will look at infectious disease research papers, while others will feature Q&As with different LCID PIs and researchers.
In our inaugural blog post, we spoke to Dr. Poulami Das, researcher in the Dudley Lab on her work which is focused on understanding ways that viruses disrupt cellular function. This Blog is in a Q&A format.
Q1: Welcome to the Blog, Dr. Das. You study a process called viral retrotranslocation. Would you tell the reader what that is?
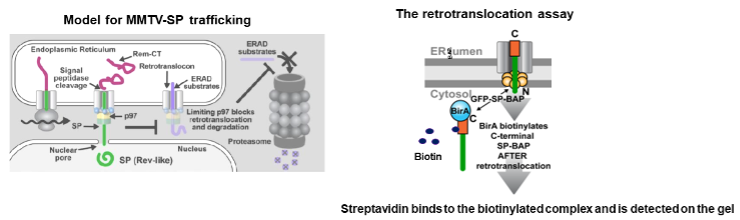
A: Thanks! The literal meaning of Retrotranslocation is ‘opposite of translocation.’ So, what is translocation then? In higher organisms, after a protein is synthesized by the ribosome, they need to be sent to their specific compartments within the cell where they can perform their destined functions. This process of sorting the proteins occurs in a part of the cell called the Endoplasmic Reticulum (ER). In mammalian cells, it occurs at the same time as the proteins are synthesized or “co-translationally.” Newly made proteins (mostly secretory and membrane proteins) use a complex pore in the ER called the translocon for entrance. This process is known as ‘translocation.’ However, before embarking on their sorting pathways, all the proteins must pass the quality control step in the ER to ensure that only completely synthesized and properly folded/functional proteins are being distributed. The improperly made proteins are sent back from the ER to the cytoplasm most likely using the same translocon system to be destroyed and degraded by another cellular component called the proteasome. Since here the translocation occurs in reverse, this process is commonly known as retrotranslocation or dislocation and the entire mechanism of getting rid of misfolded/misassembled proteins by the ER is known as ER-associated degradation (ERAD).
Q2: At what point in the viral life cycle does this process matter, and why is it interesting to you?
A: This process has been reported in eukaryotes, but several viruses exploit it in different ways for their benefit and to increase the efficacy of infection. Some viruses such as Human Immuno-deficiency virus (HIV-1) and some herpesviruses have special proteins that causes critical antiviral host proteins to undergo retrotranslocation and degradation by the proteasome. Mouse Mammary Tumor Virus (MMTV), the virus our lab studies, synthesizes a specialized signal peptide (SP) that uses retrotranslocation to move from the ER membrane to the cytosol. The SP then is imported into the nucleus where it binds to viral RNA for export to the cytosol. This process eventually leads to the synthesis of more viral particles. Besides viruses, certain bacterial and viral toxins also use the retrotranslocation process to cross the ER membrane. It’s just amazing and mind boggling to study how a microorganism can utilize one of the key quality control processes of our body for its own benefit.
Q3: Why is there a need for designing specific assays to study this system?
A: Despite being a critical process and its active involvement in pathogen invasion, there are several key aspects of retrotranslocation that are largely unknown. The first and foremost requirement in studying this is a well-defined in vitro retrotranslocation system/assay that can identify the role of different components and used to quantitate the amount of the protein/s that is being retrotranslocated.
Q4: What about your approach was unique/different?
A: There are some approaches available for studying the retrotranslocation process. But the biotinylation based assay that we have designed to study SP retrotranslocation provides a simplified yet quantitative measure of the process. The assay is based on the use of a small biochemical tag called biotin. What we do is introduce a small tag called Biotin Acceptor Peptide (BAP) tag that is engineered into the protein of interest (POI). Biotinylation is triggered by a bacterial ligase (BirA) that is localized only in the cytoplasm of the cell. So, this enzyme can biotinylate the BAP tag in the POI ONLY and ONLY IF the POI undergoes retrotranslocation and faces the cytosol. The biotinylated protein is then captured by conjugation with streptavidin which causes an increase in the mass of the POI by about ~50 kDa. In protein biochemistry, biotin-streptavdin interaction is known for its specificity and is one of the strongest.
Q5: What was the biggest technical challenge that you overcame in the completion of this project or even in the paper that came from the project?
A: A similar type of assay has been developed for several cellular proteins that are subject to retrotranslocation and degradation by the proteasome. However, this was the first time that this approach was utilized for a viral protein. We had some indirect evidence to prove that SP undergoes retrotranslocation from the ER to the cytosol prior to its entry into the nucleus. However, a direct assay was needed to quantitate SP retrotranslocation and study other factors involved in this process. When I started, the major challenge was engineering the tag at the right place since SP is made as a part of a viral protein called Rem, but SP is cleaved by a cellular protease. This was an important consideration because if I placed the tag at a position where it gets cleaved off, our assay was not going to work. An additional engineering detail was that we needed to place the tag in a region that was inside the ER. Another technical challenge was to work out appropriate controls for the assay because we wanted to be absolutely sure that what we were measuring was actually SP retrotranslocation. So, we checked whether manipulating the location of the BAP tag would allow an ER-specific ligase to biotinylate the cytosolic tag.
Q6: What is your favorite piece of data from this study, and why?
A: My favorite piece of data would be identifying the regions of SP that is absolutely required for its retrotranslocation as well as interaction with p97/VCP. VCP is a key cellular component that provides the energy required for the process of retrotranslocation via ATP hydrolysis. Any change in this region had an adverse effect on retrotranslocation. We were thrilled to find out via protein modeling that changes in amino acid residues in this region affects the conformation of SP and its ability to dimerize/multimerize.
Q7: What is your favorite assay or technique in this study and why?
A: The retrotranslocation assay itself is my favorite because I was able to show directly that SP undergoes the retrotranslocation process but evades degradation by the proteasome. Previously we were doing assays with this viral protein that provided indirect evidence of the amazing journey of SP in cellular trafficking. This assay provided not only a direct proof but also a quantitative estimation for SP retrotranslocation to the cytosol.
Q8: Is there anything else that you would like for the reader to know about your work?
A: I am still working on several aspects of the Mouse Mammary Tumor virus full length Rem. Just like the cleavage product SP, its precursor protein Rem protein follows a unique trafficking and has immunosuppressive function. The human genome has multiple copies of a defective virus known as human endogenous retrovirus type K (HERV-K), which is related to MMTV. Our studies are being extended to the HERV-K protein Rec which is very similar to Rem in its structure. We have initial evidence to suggest that SP and Rec use similar trafficking with potential consequences for protein quality control and disease.
For more information, please refer to the article:
P Das, WK Xu, AKS Gautam, MM Lozano, JP Dudley. A Retrotranslocation Assay That Predicts Defective VCP/p97-Mediated Trafficking of a Retroviral Signal Peptide. mBio. 2022;13(1):e0295321. January 4, 2022.